Add your promotional text...

Advancing Trypanosoma Parasite Research Through High-Resolution and Big-Data Methodologies
VAPPER
Variant Antigen Profiling for livestock African trypanosomes
Natural selection explains why all living things evolve to improve and pathogens are no exception. In the evolutionary arms race, trypanosomes are winning. These organisms have evolved to infect virtually all animal species on Earth. Some have done so harmlessly, others developed pathogenicity. If on one hand pathogenicity allows for higher parasite load, thus increasing transmission potential, on the other, it triggers the host’s immune response, forcing the parasite to find ways to evade or modulate it.
VAPPER
One of the most successful immune evasion strategies is antigenic variation, which Salivarian trypanosomes excel at. The parasite’s surface is coated by a single, highly immunogenic protein, so the immune system develops a strong antibody response against it. But, when it switches randomly from 2000 genes, and the genetic pool keeps increasing due to extreme recombination (Marcello & Barry 2007, Mugnier et al 2015, Jayaraman et al 2019), controlling infection becomes impossible. Therefore, understanding how variant antigens relate to each other, how diversity is endlessly generated, and how to disrupt it, is essential to control disease. Previous research showed that trypanosome variant antigens, named variant surface glycoproteins (VSGs), have evolved by distinct evolutionary mechanisms in different species (Jackson et al, 2012). For example, T. brucei VSGs evolve rapidly from a combination of multiple genetic recombination strategies and few constraints, but T. congolense VSG diversity generation seemed limited to closely-related genes. By sequencing the genomes of 69 field isolates and transcriptomes of parasites from the vector and the host, we showed evidence for purifying selection amongst T. congolense VSG lineages (Silva Pereira et al, 2018) and proved that recombination does not drive T. vivax VSG diversity (Silva Pereira et al, 2020). These previously unknown “rules” have brought hope for prophylaxis because they uncover conserved targets that can be explored in vaccine development.
Can we automate variant antigen profiling?
We developed VAPPER, the first tool for the systematic analysis of VSG diversity across strains and during infections (Silva Pereira et al, 2019). It relies on universal protein motifs and/or gene mapping to characterize differences in antigenic repertoires between strains, hosts, and conditions, which may be the starting point to build a real understanding of the association between parasite genotypes and outcomes of trypanosomiasis. Antigen profiling has been extensively explored by virologists. For example, it improved the prediction of antigenic drift in influenza A (McHardy et al, 2009), required for the efficacy of the seasonal flu vaccine. In HIV research, envelope glycoprotein profiling preceded the formulation of a multivalent subunit vaccine (McCutchan et al, 1996). In parasitology, expression of specific Plasmodium var gene lineages underlie severe malaria (Chen et al, 2011; Wang et al, 2012). However, the complexity of parasite genomes, and in particular the size and expansion of parasite-specific gene families, has prevented the development of a method that accommodates strain variation. This is especially relevant considering there are roughly 33x more VSGs in a trypanosome cell than var genes in Plasmodium, even though the trypanosome genome is only 2x bigger.
Are variant antigens functionally different?
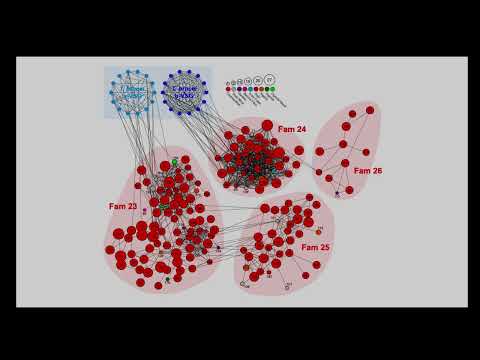
The T. congolense VAPPER was feasible because its VSG repertoire is not subject to random drift across the population. VSGs segregate into 15 stable lineages and their structured composition is a fixed feature of the species. Taking the example of T. brucei or other multi-copy gene families, functional redundancy would likely be associated with increased VSG structural diversity accompanied by fluctuations in size by random gene birth-and-death processes. Instead, in T. congolense, the relative proportions of each lineage remain constant across the population, suggesting each lineage may have a specific and essential function. These discoveries suggest that VSGs may play important role besides antigenic variation (Silva Pereira et al, 2022).
Is VSG expression in vivo structured?
When T. congolense is in its vector, the tsetse fly, VSGs are not arbitrarily expressed from the genome. The transcriptomic VSG profiles are consistently distinct from the genomic repertoire, but they are non-random, reproducible, and do not reset upon a new transmission cycle (Silva Pereira et al, 2018), contrasting with other pathogens employing antigenic variation.
We have also discovered that in the mammalian host, T. vivax VSG expression is not random either. Instead, we see co-dominant VSGs expressed across successive peaks of parasitaemia, in a partially reproducible order. Besides, this reproducibility has a strong phylogenetic signal because related lineages are expressed simultaneously or consecutively throughout infection, and lineages expressed at a given peak regularly comprise multiple distinct, but closely related, transcripts, even though their genes rarely share genomic loci (Silva Pereira et al, 2020).
Where are VSGs expressed from?
T. brucei VSGs are expressed from specialised peri-telomeric regions, called bloodstream expression sites (BES), but for T. congolense and T. vivax such knowledge is lacking. Using long-read genome sequencing, we have discovered a conserved peri-telomeric region associated with T. congolense VSG loci (Abbas et al, 2018). Despite little sequence homology, the structural similarities between these regions and the T. brucei BES are obvious, suggesting functional equivalence. Sequence divergence points towards a shared peri-telomeric BES in their last common ancestor, that has since diverged independently in each species, which might have affected the mechanism of antigenic variation.
So what?
This work showed that, whilst there may be a common phenotype of antigenic variation, the mechanisms of antigenic switching and diversification are different. In the case of trypanosomes, the large species-specific variability likely had a considerable impact on VSG expression patterns, transmission, disease progression, and virulence. Clearly, parasite adaptation strategies rely heavily on interactions with the vector, host, and environment. Yet, they may also rely on the degree of VSG structuring and expression hierarchies, as they are the most noticeable players in the host-trypanosome interaction.
VAPPER allows these hypotheses to be tested rapidly and accurately, using data from natural infections. Ultimately, the ability to take research from the lab back to the field is extraordinary. It facilitates the integration of local knowledge with cutting-edge methodological advances; it brings awareness to where the problem is; and it promotes research that is focused on sustainable solutions to control disease and improve animal welfare and economic development. As we persevere and find novel ways to tackle previously impossible tasks, such as a vaccine for African trypanosomiasis, we might well begin to win the fight against infectious diseases.